



Next: Analytical Gel to Check
Up: Create additional pBR322 vector
Previous: Miniprep to extract

Materials:

Methods:
- assuming
 DNA in
DNA in  TE buffer (
TE buffer (  because added the gel loading dye already by mistake), set
up a digest for pBR322 cut with EcoRI in a 1.5ml eppendorf tube,
by first pipetting the following:
because added the gel loading dye already by mistake), set
up a digest for pBR322 cut with EcoRI in a 1.5ml eppendorf tube,
by first pipetting the following:
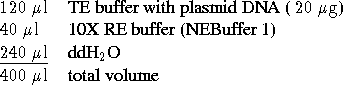
- Add
 restriction nuclease EcoRI
restriction nuclease EcoRI
 and mix the contents well by gently tapping the bottom of the tube
with finger (avoid bubbles). There is one EcoRI cut site on pBR322.
and mix the contents well by gently tapping the bottom of the tube
with finger (avoid bubbles). There is one EcoRI cut site on pBR322.
- optinally: briefly spin down the contents of the tube in centrifuge.
- incubate for 1.0 - 1.5 hours

- inactivate the restriction enzyme with 10 min
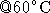
Results: ( 1/27/99)
Performed the digest on the pBR322 miniprepre sample from previous
step. Because added gel loading dye by accident, used twich as much
restriction nuclease EcoRI than needed. Heat inactivated for 15 min
 because also inactivated HindIII at same time.
because also inactivated HindIII at same time.

Ron Weiss
Wed Feb 10 15:48:08 EST 1999
![]()

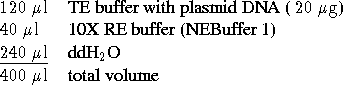
![]() because also inactivated HindIII at same time.
because also inactivated HindIII at same time.
![]()