 |
Research in the Tidor Lab
| |
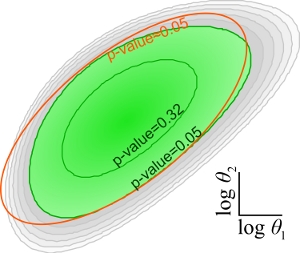
Quantifying Uncertainty in Biological Network Models
When constructing a computational model of a biological network, there is not just one model that is consistent with the available data, but many models with different parameters and different topologies. Quantifying the uncertainty in the topology and parameters is a necessary step toward quantifying the uncertainty in the predictions of the model. If the uncertainty is undesirably large, it will be necessary to collect additional data in order to reduce the uncertainty. We develop methods to quickly approximate the uncertainty in the model and to predict which experiments will reduce the uncertainty the most. ...
|
|
| |
|
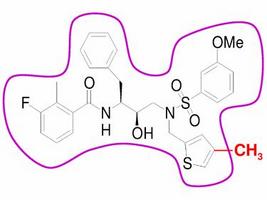
Molecular Design of Drug Resistance Resistant Inhibitors
AIDS is one of the major killers in the modern world, but despite the many treatments that have been developed, the HIV virus continues to spread. One of the main problems in designing drugs to combat HIV is the potential for the virus to develop resistance via mutation. This is a problem that affects drugs targeted at many organisms and is likely to become more of a problem with increased use of drugs to target viral and bacterial infection ...
|
| |
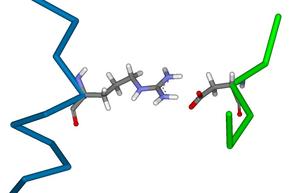
Creation of a Biological Calcium Sensor by the Rational Redesign of Calmodulin
Calcium is the most abundant mineral in the human body and serves many important biological functions. Within the mammalian brain, the control of calcium-ion flow serves as a basis for neuronal communication and is important in both learning and memory. Calmodulin is a calcium-binding protein that is found in many different cell types and in many different cellular locations. It undergoes a large conformational change upon binding calcium, allowing it to perform its downstream functions ...
|
|
| |

|
Understanding Oscillatory Biological Systems using Dynamic Optimization Techniques
Numerous examples show that oscillatory systems play an important role in biology. Some biological processes are clearly periodic, e.g. the cell cycle (1). In other systems, oscillations are used to convey function in more subtle ways. An example is the cAMP transients and Ca2+ spikes in spinal neurons, as described in reference (2). The context chosen for the present study is the circadian clock system found in many different organisms, from unicellular ones (3) to mammals (4) ...
|
| |
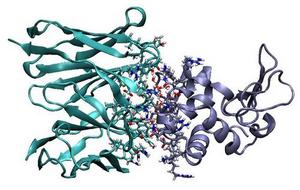
Antibody Affinity Maturation Using Computational Protein Design
The biochemical process of two proteins in solution forming a single bound complex is a basic molecular event in biology with relevance to the study of biological systems and the development of treatments for disease. The strength, or affinity, of a binding reaction is of critical importance for drug design. The aim of this project is to develop and apply computational protein design tools for engineering high-affinity protein interactions ...
|
|
| |
|
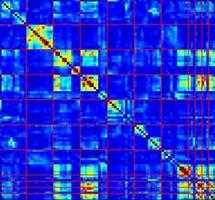
Information Theoretic Approaches to Identifying Informative Subsets of Biological Data
Understanding how biological systems process and respond to environmental cues requires the simultaneous examination of many different species in a multivariate fashion. High-throughput data collection methods offer a great deal of promise in offering this type of multivariate data, however, analyzing these high-dimensional datasets to identify which of the measured species are most important in mediating particular outcomes is still a challenging task ...
|
| |
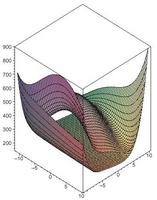
Dynamic Optimization as a Method to Probe Biological Networks and Their Behavior
The emerging field of systems biology includes a strong focus on the network perspective of biological systems. In this work we explore the development and application of tools that can be used to discover structure--function relationships in biological networks, to discover design principles and to design synthetic studies of new and modified networks ...
|
|
| |
|
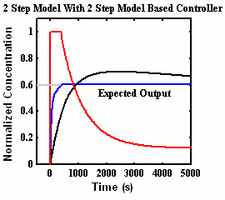
Using Optimal Time Varying Inputs to Probe Biochemical Signaling Networks
Biological signaling networks consist of complex sets of biomolecular interactions that process extracellular signals, such as physical or chemical stimulation, and affect intracellular changes such as protein modifications or changes in gene expression. Complexity in these networks emerges from a variety of factors, including the large number of interacting components, spatial compartmentalization, and feedback loops ...
|
| |
|
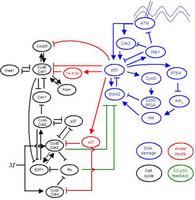
Decoding the Mechanism of Oscillations in Feedback-Connected Biological Networks
p53 is a transcription factor involved in cellular stress response, and plays a role in directing the programs of cell cycle arrest, DNA damage repair, and apoptosis. p53 protein levels are seen to undergo a series of pulses after ionizing radiation induced DNA damage. The signaling network associated with this response is complex, consisting of a number of feedback loops that both positively and negatively regulate p53 levels ...
|
| |
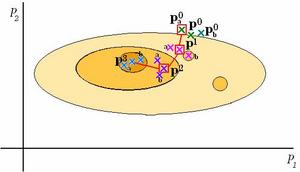
Parameterizing Biological Networks
As biological systems are being increasingly investigated from the networks point of view, there is an escalated demand for computational models that quantitatively characterize those systems. As is the case with modeling any system, accurate and precise models that can both represent and predict their respective biological systems' behaviors are desired. An essential task in building such a model involves effective calibration of the parameters that define the model ...
|
|
| |

|
Computational Analysis and Prediction of Phosphopeptide Binding Sites
A machine-learning technique has been developed for the purpose of analyzing the properties of phosphopeptide binding sites on the surface of proteins, and was applied predictively to the surface of a pair of phosphopeptide binding domains for which the ligand binding sites were not crystallographically determined (5). This technique focuses on the local chemical and physical properties of extremely small portions of the protein surface ...
|
Accessibility
|
 |
|



