![]()
Materials:
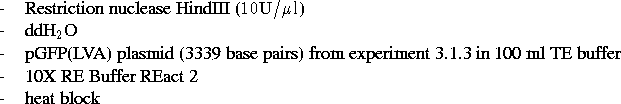
Methods:
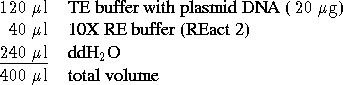
Results: ( 1/27/99)
Performed the digest on the two pGFP(LVA) samples from previous step.
Because added gel loading dye by accident, used twice as much
restriction nuclease HindIII than needed. Incubated digestion
reaction for 1 hour. Performed an analytical gel (in the small cast,
using 15 ml for each well). Figure 3.1
shows the results of an analytical gel elecrophoresis on the
restriction digest samples pGFP(LVA) cut with HindIII and pBR322 cut
with EcoRI. The figure shows that for sample 1, pGFP(LVA) appears to
have been digested properly, leaving two segments (presumably, a long
one, and the GFP(LVA) insert). There is no sign of DNA for sample 2,
leading me to believe that the miniprep went bad. Also, stored
restriction digests ![]() , instead of
, instead of ![]() . Could this have made a difference?
. Could this have made a difference?
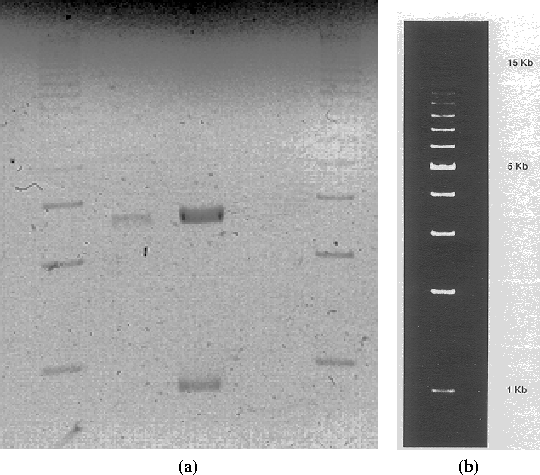
Figure 3.1: (a) Analytical gel electrophoresis (1.5 hours, ![]() ) on restriction digest samples. Lanes have: 1 = 1kb ladder, 2 =
pBR322 cut with EcoRI sample, 3 = pGFP(LVA) sample
) on restriction digest samples. Lanes have: 1 = 1kb ladder, 2 =
pBR322 cut with EcoRI sample, 3 = pGFP(LVA) sample ![]() cut with
HindIII, 4 = pGFP(LVA) sample
cut with
HindIII, 4 = pGFP(LVA) sample ![]() cut with HindIII, 5 = 1kb ladder.
(b) 1 kb ladder, not shown to scale with other figure.
cut with HindIII, 5 = 1kb ladder.
(b) 1 kb ladder, not shown to scale with other figure.
![]()