Diagnosing cardiovascular disorders requires determining the mechanisms producing the findings as well as determining the primary causes. For example, finding the appropriate therapy for a patient experiencing pulmonary congestion after a myocardial infarction requires knowing whether the mechanisms producing the congestion are primarily systolic or diastolic. To aid the physician in the diagnosis and treatment of various forms of cardiac dysfunction[1], we have developed a cardiovascular diagnostic reasoning model which represents the causal physiologic mechanisms that lead to the observed clinical findings. The model consists of nodes that represent pathophysiologic causes, therapies, and qualitative states of the physiologic parameters, the findings (clinical history, physical exam findings, and test results), and probabilistic links between the nodes. Each node can be true (the parameter is in that state), false, or unknown. Each link represents the probability that a true cause state will produce a true effect state. The diagnosis problem then is to find the most probable combinations of physiologic states that account for the findings. This paper discusses the method we have developed to handle this problem and discusses the effectiveness of the solution.
Many programs have been developed to assist in the diagnosis of medical disorders, but the emphasis has been on finding primary causes rather than mechanisms. Even current programs such as DXplain[2] only use associations between diseases and findings. The approach most similar to ours was that taken in CASNET[3], but it differs from ours in two important ways. First, their causal network primarily represents the progress of the disease and so does not include therapies and feedback loops. Second, they use a mechanism for producing diagnoses that locally determines the truth of each node rather than globally finding the combinations of nodes that best account for the findings.
One approach to diagnosis in the cardiovascular domain that does account for feedback loops is that of Widman[4]. However, his system uses simulation for determining what findings a disease or combination of diseases will produce. Therefore the diagnosis requires simulating diseases to find the ones that might produce the observed findings.
The program we are developing assists in the diagnosis of diseases that cause symptoms of heart failure at a level of detail sufficient to assist in the management of the hemodynamic dysfunctions. Thus, the program covers diseases of the cardiac muscle, valves, pericardium, as well as diseases of other organ systems as they affect or mimic effects on cardiac hemodynamics. The nodes in the knowledge base network represent causes or physiologic states. The causes have a prior probability without being caused by another node within the model. Because of the variation of these probabilities with age, sex, and other factors, we have adjustments for such dependencies in the model. For example, the node for anemia (Figure 1) sets the probability of anemia without other causes to 0.04 if age is less than 70 and 0.08 if age is greater. If age is unknown, it uses 0.05.

The physiologic state nodes have some probability of being caused by another node within the model. In the anemia example, 30%of the time chronic renal insufficiency will cause anemia.
Findings are the symptoms, physical examination results, and laboratory test results known about the patient, and are the terminal nodes of the model. In the example, anemia produces the symptom dyspnea on exertion (DOE) about 50%of the time. The probability of anemia showing up on a complete blood count (CBC) is 1.0. The knowledge base also has the false positive rates for the various findings. For DOE the knowledge base contains the statement that 70%of the time DOE is caused by nodes in the model and therefore 30%of the time DOE needs no explanation. An anemic indication on a CBC is always caused by anemia.
The resulting network of binary nodes and probabilities resembles a Bayesian belief network as defined by Pearl[5]. However, we do not require the network to be acyclic or have only single paths between nodes. Since these two restrictions are needed to make the exact calculation of node probabilities and composite hypotheses tractable in any network with more than about twenty nodes, we have developed heuristic methods to determine hypotheses from a set of findings. These methods are based on an observation about the nature of hypotheses. A hypothesis is a set of nodes in the model accounting for all of the findings and accounting for any nodes in the hypothesis that are not themselves primary. In any hypothesis each finding is explained by one or more paths from a primary cause to the finding - usually a single path. Thus, one way to determine a hypothesis is to find a set of causal pathways that account for all of the findings. This can be done by selecting a minimal set of causes from which there are causal pathways to all of the findings, taking the them one by one, determining the most probable path from a cause or node in the emerging hypothesis to that finding, and adding the path to the hypothesis. Our program uses this general algorithm, with some enhancements for optimization and heuristics for picking good sets of possible causes and ordering the findings.
This algorithm relies heavily on the ability to identify the causal pathways to each finding and to select pathways with high probability. Since determining the causal pathways is computationally intensive, we have precomputed the pathways in the model. This is done on initialization and is then available to the diagnosis algorithm. In the current model there are almost 57,000 causal pathways at the node level (not counting multiple findings from a pathway). Each pathway has the probability computed from the links that make it up. Since many of the probabilities are dependent on the facts about the patient, these must be recomputed for each case. Still, this has proven to be an effective way of searching for hypotheses.
The diagnostic process starts by identifying the abnormal findings provided by the input. These findings drive the diagnosis. The program will account for each finding unless the probability that it might be caused by something outside the model (irrelevant diseases or normal variation) is higher than the probability that it is caused by the hypothesis. The findings are first used to determine nodes that are definitely true or false. For example, if a node (such as anemia) always produces a finding (anemia on CBC) but the finding is absent, then node is false. If only one node can account for a finding which is present, the node must be true. This process usually produces a few node values. The next step is to identify primary causes which account for a large number of the findings. This is done examining the causal chains to each finding to list its possible causes. Causes which could account for large numbers of findings are used to generate hypotheses. Because one cause may not account for every finding, additional causes are found for each of the major causes to produce one or more sets of covering causes.
Given a set of causes and the findings, the program must find the likely intermediate nodes that specify the mechanism. This is done by sorting the findings by their probability of being caused by one of the causes, then using each finding to fill out the hypothesis. When each finding is considered, there is a partial hypothesis from which to start. The program finds the most likely path from the hypothesis nodes to the finding. One of the nodes already in the hypothesis may account for the finding, an already selected intermediate node may provide a short path to the finding, or the best path may start at one of the causes. It is also possible that the probability of the finding being caused by something outside the model is higher than any explanation within the model. In that case, the finding is left without a cause.
The hypotheses generated are ranked by their total probabilities. That is, the program computes the probability of each hypothesis node and finding using the other nodes in the hypothesis. The total probability is the product of these individual probabilities. To determine the probability of the hypothesis given the findings, it would be necessary to divide this number by the probability of the findings. Since we only need to compare different hypotheses having the same findings, it is unnecessary to find the probability of the findings. The program gives as the differential diagnosis the hypothesis with the highest probability and any other hypothesis with a probability higher than 1%of that probability. This process may produce a single hypothesis or a dozen, but usually two or three. Each hypothesis in the differential diagnosis can be displayed for the user as a graph of the nodes in the hypothesis with the links between them, supplemented by information about the probabilities along the paths.
Consider a patient entered by the user. The summary given by the program is as follows:
From this description the program determines twenty findings to be explained. These drive the algorithm to produce a differential diagnosis. In this case the differential contains the following five hypotheses with their relative probabilities:
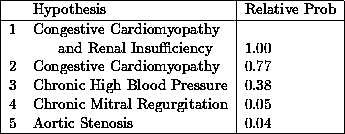
These are all reasonable hypotheses. The first two differ by including renal insufficiency as a better explanation of the renal findings. The third is essentially hypertensive heart disease as a cause for cardiomyopathy. The last two hypotheses are important because the murmur might indicate valve disease that could benefit from surgery.

The computer display of the highest probability hypothesis is reproduced in Figure 2. This figure shows the causal explanation that the program is proposing as the most likely explanation of the patient's findings. The findings are shown in lower case and the nodes in upper case. The four nodes and one finding that are considered primary or diagnostic are printed in bold. The numbers in parentheses following the names indicate the a priori probability that the item is primary. The arrows indicate the most important causal relationships. (Ones with low probability are left out of the display if there is a higher probability link in the hypothesis. For instance, digitalis can also cause nausea/vomiting, but renal insufficiency is a more likely cause.) The numbers on the arrows are the probability that the cause will produce the effect. Links with a W+ increase the probability of the effect but can not cause it alone. Links with P- decrease the probability of the effect. The probabilities from nodes to findings have been left out to keep the display readable.
This example illustrates several points about the generation of causal hypotheses. First, the program generates hypotheses containing multiple causes. In this case congestive cardiomyopathy with renal insufficiency will account for the findings. It happens that congestive cardiomyopathy alone will also account for the findings (the second hypothesis) with only slightly lower probability. In the actual case, a primary concern of the physicians involved was whether the rising BUN was prerenal or renal; these two hypotheses capture that problem. The program will also assign multiple causes for a single node when appropriate. For example, the high left atrial pressure is linked to both high venous volume and low cardiac output. Also, the cause of the high BUN does not have to be either renal insufficiency or low renal perfusion, it can be both.
Second, the program does not have to find causes for all of the findings. In this case, systolic ejection murmurs are so commonly heard without indicating a significant disorder that it is better to leave the murmur unexplained than generate a possible mechanism consistent with the rest of the hypothesis. On the other hand, the two valvular hypotheses use the murmur as key evidence.
Third, the program recognizes that some findings can be iatrogenic. A possible explanation for the runs of ventricular tachycardia in the example is a toxic reaction to the digitalis. In the actual case, the patient had been taking double the prescribed dosage and was indeed toxic.
Finally, the mechanisms proposed in the display are a rich source of information for management as well as diagnosis. Looking at the causal chains, places where the patient might be responsive to therapy include decreasing the blood volume with more diuretics, promoting left ventricular emptying by decreasing the vascular resistance with a vasodilator, increasing the low systolic function with stronger inotropic agents, and stopping the digitalis to eliminate the toxic effects. None of these actions are directed at the two primary causes in the hypothesis since those are difficult to treat. Rather, the appropriate therapies take advantage of the mechanisms and attempt to break the causal chains.
Over the past year we have collected and examined the program's performance in 42 cases selected with the intention of challenging the diagnostic strategy and improving the knowledge base. Of these, the program produced an acceptable differential with justifiable hypotheses in 31 cases. In 5 cases the hypotheses were almost right but had parts of the mechanisms that were inappropriate. In the remaining 6 cases the best hypothesis was missed. We have improved the algorithm and the knowledge base during this time, so this is not an evaluation, but it is still a reasonable measure of the potential competence of the method in its present form.
There are two main reasons why the program does poorly on some cases: lack of mechanisms to use the time relations among causes and findings, and inadequate use of severity. A patient who had a myocardial infarct and was treated with nitroglycerin a few hours later illustrates the time problem. After treatment the patient still had signs of pulmonary congestion but the pulmonary capillary wedge pressure had normalized. The program proposed pneumonia to explain the pulmonary congestion because cardiac mechanisms required a high wedge pressure. Even though nitroglycerin had reduced the high wedge pressure to normal, it was still the proper mechanism to explain the lung findings since signs of pulmonary congestion may take several hours to disappear. Quantifying severity became an issue in a few cases where patients had very low blood pressure. The program suggested dehydration from fever or diuretics as causes, but although they can lower the blood volume and hence the blood pressure in a patient, they are not the most likely mechanism for a severely reduced blood pressure of 80/50.
To address the time issue we are extending earlier work on the logical relationships among causes and effects over time[6] to handle probabilistic relationships. The challenge is to extend the model to represent multiple causal chains with the same physiologic nodes at different times and still be able to precompute causal chains to keep the program computationally usable in an interactive environment. Severity could be handled either by identifying more states for the parameters (e.g., very low) or by adjusting the probabilities between cause and effect for the severity of findings. That is, findings that require a certain cause severity would constrain the node and change the associated probabilities. The last method looks most promising but the details remain to be worked out.
The algorithm for generating hypotheses does not always find the best hypothesis and we have found several situations where it misses better hypotheses:
This method has proven to be useful in generating differential diagnoses for difficult cases requiring a physiologic approach to reasoning. The primary advantage of the method is that it justifies the hypotheses with proposed mechanisms, providing the user with the necessary information to evaluate the hypothesis and consider possible therapies. Even if the program does not list the best hypothesis, the hypotheses generated by the program can be an effective way of helping the user organize the information and remind him of better hypotheses.
This document was generated using the LaTeX2HTML translator Version 0.5.3 (Wed Jan 26 1994) Copyright © 1993, Nikos Drakos, Computer Based Learning Unit, University of Leeds.
The command line arguments were:
latex2html -split 0 web/papers/cinc88.tex.
The translation was initiated by wjl@MEDG.lcs.mit.edu on Wed Aug 31 15:36:20 EDT 1994