Web Interface for the Heart Disease Program
William J. Long1, Hamish Fraser1, 2, Shapur Naimi2
1MIT Lab for Computer Science, Cambridge, MA,
2
Tufts-New England Medical Center, Boston, MA This paper was presented at the American Medical Informatics Association Conference, November 1996 and appears on pp. 762-767 of the proceedings.The task of making a large complex diagnostic program available to a broad audience of physicians has become more feasible with the ubiquitous accessibility of the client-server architecture of the World Wide Web. This paper describes the design and implementation of a Web interface for the Heart Disease Program (HDP). The client-server architecture imposes a number of requirements on the program. The graphical capabilities of the Web enable a number of enhancements to the program but also cause some limitations. Our initial experience with physicians using the HDP through the Web interface has been positive and we are now conducting an evaluation of the HDP using this form of access.
INTRODUCTION
Over the past dozen years we have developed the Heart Disease Program(HDP)[1,2] to help physicians diagnose the range of conditions that cause hemodynamic dysfunction. The physician enters a patient description including the history, current therapies, physical examination findings, and laboratory test results. The program then uses a pseudo-Bayesian probability network representing the causal relationships among the physiologic states to generate a differential list of hypotheses to account for the findings. The use of a novel heuristic reasoning strategy incorporating severity and time constraints permits a more accurate representation of the behavior of the cardiovascular system than the standard Bayesian belief network. The hypotheses are ordered by likelihood and those below a threshold are removed from the list.
The resulting differential diagnoses can be displayed to the user in a variety of forms, from a causal graph to an outline summary. Since the program does considerable reasoning about the findings and relationships, it can provide several kinds of information to justify the hypotheses. The program has undergone a first stage of testing using 26 cases of heart disease. The diagnostic summaries were assessed by 5 cardiologists and the system was considered to be sufficiently accurate to warrant further testing.[1]
The HDP is written in COMMON LISP and runs on a Sun Sparkstation 20 where the LISP image occupies about 25 megabytes and cases take half a minute to five minutes to process. It is therefore a poor candidate for distribution in the traditional manner because the intended audience of physicians tends to utilize low to middle range PCs and the program is continuously under development to improve its accuracy and speed. For these reasons, the best way to make the program accessible to physicians is on a central server accessible over the World Wide Web[3] using a standard browser.
The existing infrastructure of the Web provides tremendous opportunities and a number of limitations that represent a challenge for the use of the Web for client-server software. This paper will describe the implementation of the Web interface for the HDP, the benefits and problems with the use of the interface as it presently stands, and the implementation of a scheme for the evaluation of the program utilizing the Web interface. Although a number of other medical projects have made creative use of Web (e.g., [4]) and some even provide interfaces to expert systems (e.g., [5]), the HDP presents particularly demanding requirements for both input and textual output.
Requirements
The challenges of providing an interface for the HDP are the range and complexity of the input data, providing the user with an appropriate summary of the conclusions, and allowing the flexibility of use that would be expected in a standalone diagnostic aid. The HDP accepts and utilizes whatever data is entered. Besides the history, symptoms, known diagnoses, therapies, and physical exam, there are a number of tests that have relevance to the diagnosis of heart disease. Some of these are frequently done, such as the EKG and chest X-ray, but others such as pulmonary angiography are infrequent but very pertinent if available. Each of the tests has a number of possible values represented in the HDP. For example besides the time of the test there are about 50 possible values for the EKG.
Many of the symptoms require detailed description to enable the program to handle them reasonably. For example, if the patient has hemoptysis (coughing up blood), it is useful to know whether the sputum is pink and frothy or blood streaked, how long this has been taking place, the approximate frequency and how recently the last episode took place. It is prohibitive to have all possible input values on a single form: the users would be overwhelmed. Even if they persevered it would be easy to get lost and miss important data entries. In entering a typical case the user clicks on 20 to 50 items out of the thousand or so possible. Thus, strategies that increase the density of the most commonly needed values were important.
Another challenge is presenting the analysis of the HDP. The program generates a differential consisting of a number of hypotheses that explain the findings. Each is a detailed causal explanation with severities and time constraints giving the program's assessment of how a set of diseases, complications, and intermediate physiologic states are likely to account for the findings. Each hypothesis contains 30 to 100 abnormal physiological states and findings with additional normal states. Thus, it is necessary to summarize the chief differential diagnosis and the other hypotheses to give the user a useful analysis of the case. Finally, users want to be able to access, modify and print cases they have previously entered. With the active case, the user may want to add or remove findings in order to perform a sensitivity analysis on the diagnosis.
Interface Design
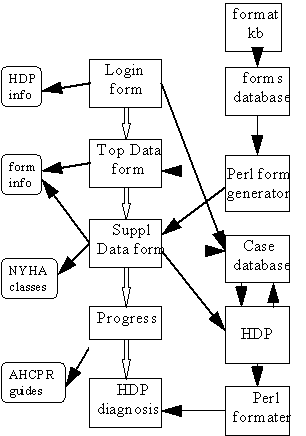
A diagram of the interface is given in figure 1. From the user's perspective, the essential features of this design are captured by the progression of documents down the center of the figure. They first see a login form, offering access to previous cases or the entering of a new case. On entering a case, there is a series of forms, each determined by the answers to the previous form. When these are completed, the case is sent to the HDP and the user is informed of the progress. When the case is completed, the differential diagnosis is returned as a document.
In addition to these primary documents, there are a few supporting documents the user may refer to along the way (on the left in the figure). The "form info" provides an overview of the structure of the forms to allow the user a preview of the lower level menus for a question. This document is provided as a separate window with many internal links to allow the user to go back and forth. Other linked documents include pertinent resources on the Web such as the AHCPR guidelines for patients with LV systolic dysfunction.

Figure 1: Block diagram of HDP interface
To allow appropriate access to past cases, follow-up when necessary, and to keep track of user status, it is necessary to determine the user's identity through the initial login form. This form also serves as an application form for new users and a mechanism for giving access to new users (with appropriate passwords).
We chose to implement the interface with an initial form covering all of the general categories and common findings followed by custom generated supplemental forms to fill in the details. An alternative would be to divide up the form into many screen sized parts. This approach was rejected to decrease the time involved in communicating with the server. Once the top level form is submitted, a second form is generated with the items entered and questions for each of the items that have further specifications or properties. This approach allows everything to be entered but has the drawback that specifying the information about a particular finding, such as dyspnea, may be split over multiple forms with other findings entered in between. However it is possible to revise part of a completed form later without losing other data. Several factors determined the architecture of the multiple forms. First, the accumulating data must be remembered with each interchange between server and client. Since identification of the user is unreliable, information must either be stored with the client as persistent information ("cookies") or must be passed back and forth as hidden values. As client storage is limited and only available in some browsers, hidden values were used here. All of the forms after login must be generated during the session to incorporate the appropriate hidden values.
Both to make the process of generating the forms more efficient and because the appropriate presentation of the data requests on the screen in HTML is quite different from the native input interface of the HDP, it was necessary to provide a database of formatting information from which the form generator could quickly put together appropriately formatted questions for each form. A static form database is inappropriate because the screens are evolving as we gain experience and the knowledge base of the HDP continues to evolve as well, making it difficult to keep the forms consistent with the HDP. Form generation is a two step process. First, a supplementary knowledge base is generated by the HDP. This contains HTML formatting templates for each input entity and provides a function that generates the form database consisting of input measures, properties, and formatting instructions. This way, the HDP is able to check the consistency of the information in the forms database against its knowledge base before use. The forms are then generated by a PERL script utilizing this file.
Once the data has been entered and is submitted to the HDP, the analysis varies from half a minute for simple cases to several minutes for a case with a many ambiguous findings. Thus, it is necessary to make the interchange very flexible. The interface uses "server-push" to keep the user informed of the progress of the analysis including an estimate of how long it will take, extrapolated from the number of hypotheses that need to be examined. If the user decides to quit and come back later, a case identifier is generated and the case is automatically included in the user's database. The HDP operates out of the case database to maintain independence from the capriciousness of the network. The CGI script stores the collected data in the database and starts the program as an independent process. The HDP then stores its progress and results in the database to be retrieved by the CGI script. The final step is for the PERL script to add the appropriate HTML formatting information to the differential diagnosis to display it on the userís screen.
Input Formatting
The presentation of the input is very important in determining the ease with which physicians can use the program. With the quantity of information in the forms, it is necessary to order it as the user would expect, concentrating data which is most commonly used, and formatting it so entries can be found quickly. The first form is the overview of the case presentation and is ordered in the same way as a clinical case summary: history, vital signs, physical examination, laboratory results, and a final section of specialized laboratory results. Concentrating the findings is accomplished by putting the general findings at the top level and the details on later forms, and by dividing categories with many possible findings into common and uncommon values. For example, one might click on anginal under chest-pain in the history section of the top form:
![]()
![]()
Once that has been submitted, a final form will ask for temporal details of the pain:

There may be three or rarely four successive forms (of decreasing length and increasing specificity) before the program is called. An example of concentrating the values is the jugular pulse:
![]()
Since test results are often normal, the top level form allows this fact to be entered rather than requiring a second form with all the possible values. Thus, EKG on the first form appears:
![]()
The test time allows the user to make the important distinction between current tests and earlier tests that may provide important information, but not reflect the present. The unknown buttons are necessary because HTML radio buttons can not be "turned off". An explicit unknown allows the user to deselect an entry. The formatting makes extensive use of tables to provide a consistent structure to the values. For example, consider part of the details of the EKG:

The check boxes here allow multiple entries per section in contrast to the radio buttons.
DIAGNOSTIC SUMMARIZATION
The diagnostic output consists of four parts: the case description, a summary of the leading hypothesis, a table comparing the various hypotheses in the differential, and a list of the abnormal findings with the states that account for them in the hypotheses. The case description is included for reference while examining the diagnosis, allowing the user to check the accuracy of their data entry, and to provide a self-contained final document. The case description is presented as a list to keep the format similar to the way it was entered. Originally, we presented it as a paragraph but that was found to be more difficult to follow.
The leading hypothesis is then summarized[6]. The summarization procedure starts with the states of nodes in the network that link findings with diagnoses. States that reflect physiological mechanism are removed, and the remainder clustered into clinically relevant states and recognized syndromes. For each state in the summary, the supporting findings are given as well as common but absent findings (as negative evidence). The diseases in the summary are presented in outline form, indicating the likely causality with primary states and those of greatest clinical significance at the top level. The differential is presented as a table with the hypotheses as columns and the states as rows. The hypotheses are ranked in descending likelihood. The states are specified in only enough detail to indicate what is different about the hypotheses. This table indicates what other, possibly important, disease states have a significant probability of being present. This has some utility for example when pulmonary embolism is a possibility, even if it is not the most likely diagnosis it is both dangerous and treatable.
The final section is essentially an inversion of the hypotheses of the differential. It lists each abnormal finding and the various ways it is accounted for in the hypotheses. This is to help of identify the findings which provide the main support for particular diseases.
DISCUSSION
During evaluations by several physicians the following benefits of this style of interface and the techniques used in this implementation were noted:
1) Predominantly mouse driven data entry speeds program use by the many clinicians who donít type well.
2) The help files and guidelines are readily accessible at the point where they are needed.
3) The wide availability of the interface on any machine with a (easily available) Web browser.
4) The ability of the development team to observe the functioning of the program and respond quickly to problems, making fixes available to all users as soon as they are made.
5)Real time scrutiny of diagnoses by cardiologists in the research group.
6)The ability to run the main program on suitable hardware.
This technology has also enabled us to conduct an evaluation of the HDP with physicians at multiple sites using a variety of hardware. To do the evaluation, we added two steps to the succession of forms in figure 1. First, after the user submits the case to the HDP, he or she is presented with an additional form asking for the physician's diagnosis (for comparison). Secondly, when the diagnosis is returned to the physician a critique form is included to allow the user to assess the accuracy and usefulness of the result. Once this is submitted, the user receives a copy of the diagnosis suitable for printing. Thus, with fairly simple additions we are able to utilize this same technology as a tool for conducting an evaluation, including such features as blinding the user to the program's diagnosis when entering their own (and visa versa).
There are also a number of drawbacks to the present system. Some of these have potential fixes as the Web technology develops.
1) The use of multiple forms: the user has to come back to previously entered information on the supplemental menus (as in the chest pain example above). It would be much better if the questions about details of a value were available immediately after the value were clicked. Using JAVA applets to generate immediate submenus should be a reasonable solution to this problem.
2) Constraints on values: Numeric values have bounds and not all combinations of categorical values are possible. At present, this is enforced either on subsequent forms or with error statements in the diagnosis results. A more immediate way of handling these constraints would be afforded by either JAVA or JAVASCRIPT.
3) Paucity of mechanisms for navigating a form: a definite limitation of the present generation of browsers. Some users wish to tab between fields and check boxes (in addition to the numerical entries) with a keystroke instead of clicking the mouse. Paging through the form is easier but still somewhat cumbersome. The scroll bar works on all browsers and some paging keys work on some browsers. Because the paging keys vary, we have included internal URLs to allow the user to move through the document. Unfortunately, internal URLs do not allow return to certain PERL generated forms, a problem with current browser design.
4)Security: this is currently difficult to enforce. We therefore avoid any identifying information more specific than date of birth in the patient record and assign our own identifiers to the cases. The rapid progress in encryption and certification should offer alternative ways to do this fairly soon.
5)Time of data entry: a major problem is the 10-20 minutes required to enter a case. This is a general problem with clinical data input: incorporation of on-line data when available should help.
CONCLUSIONS
We have developed an interface for the Heart Disease Program allowing access from the Web. The interface allows the user to enter cases in as much detail as the HDP is able to reason with. The interface uses multiple forms created on the fly whose structure is based on previous answers. It runs on a fast workstation and produces a diagnostic analysis of the case appropriately formatted as an HTML document.
The interface is acceptable to users and with simple extensions is being utilized as a tool for conducting an evaluation of the HDP.
New web-based tools such as JAVA should allow more flexible and reactive input menus. However the paradigm used here has the benefit of allowing rapid prototyping, and runs under Windows 3.1 and on virtually all modern browsers. Most importantly the forms generated are very compact taking little time to transfer, a key issue when the clinical trial involves many physicians with dial-up connections. We anticipate the program being used by physicians in assessing patients with possible heart disease, the data required at present makes it unsuitable for direct patient use.
Acknowledgments
This work was supported by the National Heart Lung and Blood Institute (R01-HL33041).
References
1. Long WJ, Naimi S, Criscitiello MG. Evaluation of a New Method for Cardiovascular Reasoning. Journal of the American Medical Informatics Association. 1994;1:127-141.
2. Long W. Medical Diagnosis Using a Probabilistic Causal Network. Applied Artificial Intelligence. 1989;3:367-83.
3. Berners-Lee TJ, Cailliau R, Groff JF, Pollerman B. World-Wide Web: The information universe. Electronic networking: Research, Applications and Policy. Westport, CT: Meckler Publishing, 1992:52-58. vol 2
4. Kohane IS, Greenspun P, Fackler J, Szolovits P. Accessing Pediatric Electronic Medical Record Systems via the World Wide Web. Pediatric Research 1995;37:139A.
5. Widman LE, Tong DA. EINTHOVEN on the World Wide Web: A tool for the Analysis of Cardiac Arrythmias. In: Gardner RM, ed. Symposium on Computer Applications in Medical Care. New Orleans, Louisiana: Hanley & Belfus, 1995:968.
6. Long WJ, Naimi S, Criscitiello MG. Summarization of Complex Causal Diagnostic Hypotheses. In: Ozbolt, JG, ed. Symposium on Computer Applications in Medical Care. Washington,DC: Hanley & Belfus, 1994:970.