



Next: Electroporate pBR322+GFP(LVA) into
Up: Transform and grow colonies
Previous: Transform and grow colonies

Partly from Sambrook, 1.68.
Materials:

Methods:
- Make a more concentrated solution of DNA fragments pBR322 and
GFP(LVA), by using
 total of the pBR322 vector and
GFP(LVA) insert with a microcon centrifugal filtration device
(according to Protocol G). Result:
total of the pBR322 vector and
GFP(LVA) insert with a microcon centrifugal filtration device
(according to Protocol G). Result: 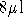 of solution. DNA fragments should be in TE (pH 7.6).
of solution. DNA fragments should be in TE (pH 7.6).
- set up tubes to anneal and ligate adapters, GFP(LVA), and pBR322
vector:
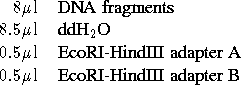
- Incubate solution
 for 5 min to melt any cohesive
termini that have reannealed. Chill the mixture to
for 5 min to melt any cohesive
termini that have reannealed. Chill the mixture to  .
.
- add the following to tube (for a total of
 ):
):

- Incubate the reactions for 4-16 hours
 (rather than
suggested 1-4 hours
(rather than
suggested 1-4 hours 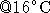 ).
).
Results: ( 2/1/99)
First, attempted to get a more concetrated form of the DNA
fragments using cetrifugal filtration. The first filtering step
seemed to work fine. However, we could not get the DNA out with 3 min
of 1,000 g. Therefore, ended up using 30 sec of 14,000 g, which
got us about 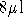 . Finally, mixed the solutions (with ligase
and ligase buffer by accident), warmed to
. Finally, mixed the solutions (with ligase
and ligase buffer by accident), warmed to 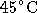 for 5 min, added
ligase and ligase buffer, and put
for 5 min, added
ligase and ligase buffer, and put  .
.

Ron Weiss
Wed Feb 10 15:48:08 EST 1999
![]()

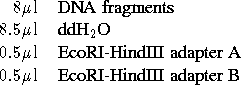
![]()
![]() . Finally, mixed the solutions (with ligase
and ligase buffer by accident), warmed to
. Finally, mixed the solutions (with ligase
and ligase buffer by accident), warmed to ![]() for 5 min, added
ligase and ligase buffer, and put
for 5 min, added
ligase and ligase buffer, and put ![]() .
.
![]()