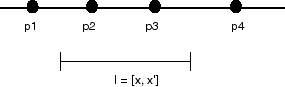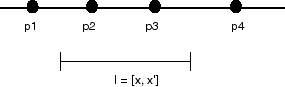1D Binary Searching: a warmup
Resources:
de Berg, chapters 5 and 10.
Searching for points on a line:
Given a list of N (unique) points on a line, locate all the points in a
given interval on that line.
Input: {p1, p2, p3, p4, ... pN}, [x, x']
Output: {pi | x <= pi <= x'}
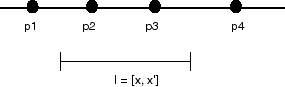
First try: Sort all the points, then scan through the points.
Output all points in the range.
This is O(N), of course we can do better!
Second try: Build a balanced binary search tree, that looks like:
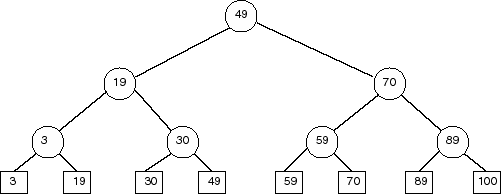
All points stored in the right subtree of a node are greater than the
splitting value stored at the node. All points in the left subtree
are less than or equal to the splitting value. To find the points in a
given range, proceed as follows:
FindPoints([x, x'], T)
if T is a leaf node, then
if x <= val(T) <=
x' then
return { val(T) }
else
return {}
end if
end if
<else T is an interior node of tree>
if x' <= val(T) then
return FindPoints([x, x'],
left(T))
else if x > val(T) then
return FindPoints([x, x'],
right(T))
else <interval spans splitting value>
return FindPoints([x,
x'], left(T)) union FindPoints([x, x'], right(T))
end if
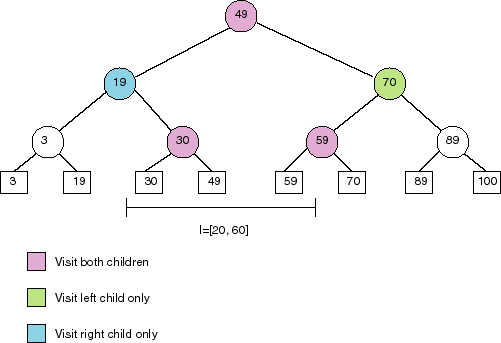
Time complexity: This search takes O(log n + k) when there are
k values in the interval, and n values in total.
How to build? There are two ways to build the tree.
-
Top down: sort all the points, then proceed as follows:
-
If there is one point only, return a leaf node with that point.
-
If there is more than one point, partition the points about the median
into two subsets, and recursively build a tree for each subset. Return
a node whose value is the median, and whose left and right children are
the two recursively constructed subtrees.
-
Bottom up: again, sort all the points.
-
Make a leaf node for each point.
-
Group the leaf nodes into pairs, under a parent node (maintain less-than/greater
than order).
-
Recursively group these nodes into pairs until there is only one node (the
root node) left.
What if the points are not unique? We just have to be careful about
our grouping. We could, for example, put more than one point into
each node, if the points' coordinates are equal.
Other variations: store the median valued points in the internal
nodes.
Searching for intervals on a line:
Given a list of N (possibly overlapping) intervals on a line, locate all
the intervals overlapping a given interval on that line.
Input: {I1, I2, I3, IN}, [x, x'] where Ii = [xi, xi']
Output: {Ii | Ii overlaps [x, x']}
If we want to find intervals which overlap with a given point x, use
the interval [x, x] as the query interval. In the following diagram, the
query interval (marked in purple) overlaps with 4 input intervals (marked
in green):
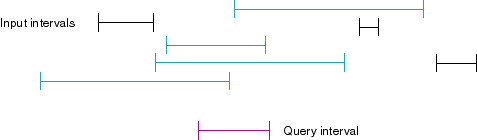
First try: stupid linear tricks....
Second try: Build an interval tree.
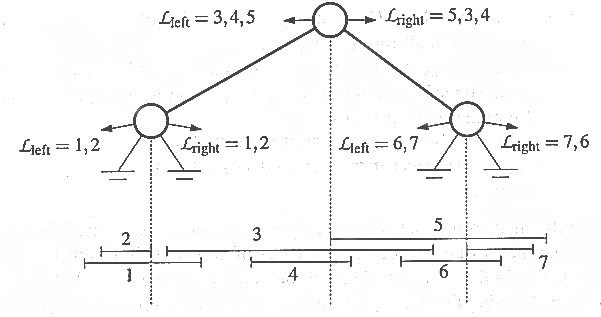
-
Partitioning values in this tree are also based on medians, but the median
values are taken from the 2N endpoints of all the intervals.
-
Left subtrees contain intervals completely to the left of the partitioning
value; right subtrees contain intervals completely to the right of the
partitioning value.
-
Intervals which span the partitioning value of a node are stored at that
node; this means we don't have to store an interval multiple times, e.g.
when there are lots of overlapping intervals.
Finding the intervals: This is just like in the point tree case.
We recurse...
If the input interval contains the partitioning
value stored at the root, then
check the input interval
against all the intervals stored at that node
else recurse on the left or right or both children,
as appropriate
Notes:
-
These trees can be constructed similarly to the point binary trees.
-
There are also variations, e.g. do we push intervals all the way down to
the leaf nodes, or do we keep them at internal nodes?
Take home idea:
Divide datasets according to some partitioning value, which lets us build
a tree structure. This gives us log n algorithms instead of linear
algorithms. Some of this issues (e.g. do we store values in interior nodes,
or in leaves? do we build bottom up or top down?) show up in the higher
dimension structures also.
[Start] [Back]
[Next]
Douglas De Couto
decouto@graphics.lcs.mit.edu
Joshua Napoli
napoli@mit.edu
Last modified: Tue Mar 17 09:55:49 1998